In Silico Study of the YAKRCFR Peptide Structure and Its Interaction with Human Peroxiredoxin-5
Abstract
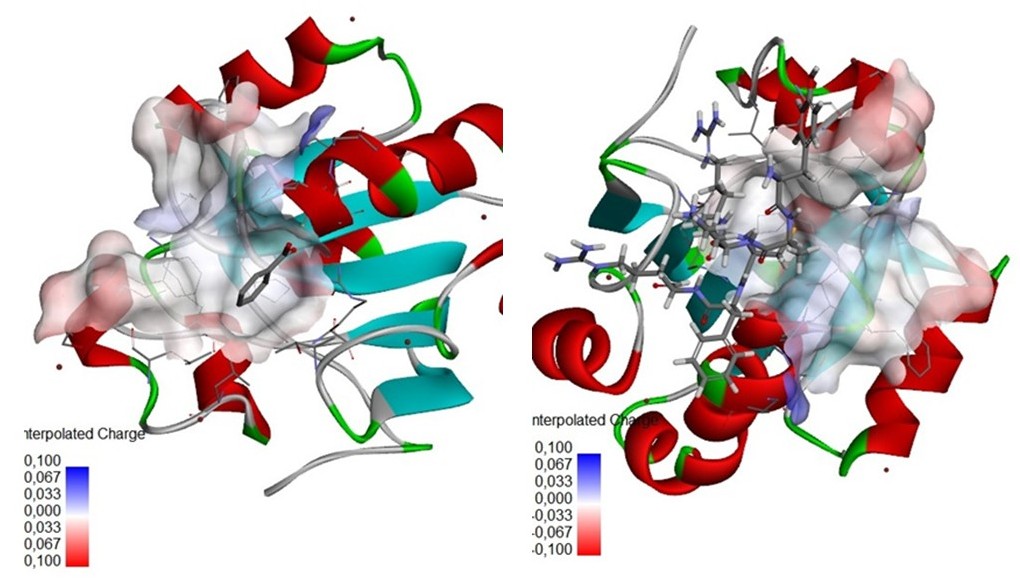
The identification of bioactive peptides with therapeutic potential is an emerging focus in drug discovery. In this study, we evaluated the structural stability and binding affinity of the oyster-derived peptide YAKRCFR through molecular modeling and docking simulations against the human peroxiredoxin receptor. Structural prediction using the PEP-FOLD4 server revealed a consistent α-helical conformation across all models, stabilized by key intramolecular hydrogen bonds and favorable sOPEP energy values. Molecular docking was validated with a root mean square deviation (RMSD) of 0.273 Å, confirming the reliability of the docking protocol. The YAKRCFR peptide exhibited a strong binding affinity with the 1HD2 receptor (ΔG = –8.1 kcal/mol), outperforming both ascorbic acid (–6.1 kcal/mol) and the native ligand (–4.862 kcal/mol). Detailed interaction analysis indicated that YAKRCFR forms stable hydrogen bonds and van der Waals interactions with critical residues such as ILE A119 and PHE A120, contributing to its thermodynamic stability and binding specificity. These findings suggest that YAKRCFR holds promise as a lead compound for further development in peptide-based therapeutic strategies, particularly for targets involving the human peroxiredoxin receptor.
Downloads
References
A. A. Aloanis, T. Herlina, A. Hardianto, and R. Maharani, “Alanine-Rich Cyclopeptides: Natural Resources, Bioactivity, Total Synthesis, and Spectroscopy Identification,” ChemistrySelect, vol. 10, no. 1, p. e202404379, 2025, doi: 10.1002/slct.202404379.
R. Maharani and M. I. Muhajir, “An overview of chemical synthesis of antiviral peptides,” Science, Engineering and Health Studies, pp. 21010003–21010003, 2021.
V. Gogineni and M. T. Hamann, “Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology,” Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1862, no. 1, pp. 81–196, Jan. 2018, doi: 10.1016/j.bbagen.2017.08.014.
R. Maharani et al., “Synthesis, Antioxidant Activity, and Structure–Activity Relationship of SCAP1 Analogues,” International Journal of Peptide Research and Therapeutics, vol. 27, no. 1, pp. 17–23, 2021.
Y. Zhu, K. Wang, X. Jia, C. Fu, H. Yu, and Y. Wang, “Antioxidant peptides, the guardian of life from oxidative stress,” Medicinal Research Reviews, vol. 44, no. 1, pp. 275–364, 2024, doi: 10.1002/med.21986.
H. Huang et al., “Isolation and characterization of antioxidant peptides from oyster (Crassostrea rivularis) protein enzymatic hydrolysates,” Food Science & Nutrition, vol. 11, no. 1, pp. 261–273, 2023, doi: 10.1002/fsn3.3058.
B. Kim, J. Park, K.-T. Chang, and D.-S. Lee, “Peroxiredoxin 5 prevents amyloid-beta oligomer-induced neuronal cell death by inhibiting ERK–Drp1-mediated mitochondrial fragmentation,” Free Radical Biology and Medicine, vol. 90, pp. 184–194, Jan. 2016, doi: 10.1016/j.freeradbiomed.2015.11.015.
H.-I. Choi, S. K. Ma, E. H. Bae, J. Lee, and S. W. Kim, “Peroxiredoxin 5 Protects TGF-β Induced Fibrosis by Inhibiting Stat3 Activation in Rat Kidney Interstitial Fibroblast Cells,” PLoS ONE, vol. 11, no. 2, p. e0149266, Feb. 2016, doi: 10.1371/journal.pone.0149266.
C. Hu et al., “Peroxiredoxin‐5 Knockdown Accelerates Pressure Overload‐Induced Cardiac Hypertrophy in Mice,” Oxidative Medicine and Cellular Longevity, vol. 2022, no. 1, p. 5067544, Jan. 2022, doi: 10.1155/2022/5067544.
J. Rey, S. Murail, S. de Vries, P. Derreumaux, and P. Tuffery, “PEP-FOLD4: a pH-dependent force field for peptide structure prediction in aqueous solution,” Nucleic Acids Research, vol. 51, no. W1, pp. W432–W437, Jul. 2023, doi: 10.1093/nar/gkad376.
A. A. Aloanis, T. Herlina, A. Hardianto, and R. Maharani, “Total Synthesis of Cyclosenegalin A,” ChemistryOpen, vol. 13, no. 12, p. e202400175, 2024, doi: 10.1002/open.202400175.
A. Nicolussi, S. D’inzeo, C. Capalbo, G. Giannini, and A. Coppa, “The role of peroxiredoxins in cancer (Review),” Molecular and Clinical Oncology, vol. 6, no. 2, pp. 139–153, Feb. 2017, doi: 10.3892/mco.2017.1129.
S.-J. Jeong, J.-G. Park, and G. T. Oh, “Peroxiredoxins as Potential Targets for Cardiovascular Disease,” Antioxidants, vol. 10, no. 8, Art. no. 8, Aug. 2021, doi: 10.3390/antiox10081244.
V. Binette, N. Mousseau, and P. Tuffery, “A Generalized Attraction-Repulsion Potential and Revisited Fragment Library Improves PEP-FOLD Peptide Structure Prediction,” J Chem Theory Comput, vol. 18, no. 4, pp. 2720–2736, Apr. 2022, doi: 10.1021/acs.jctc.1c01293.
J. Eberhardt, D. Santos-Martins, A. F. Tillack, and S. Forli, “AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings,” J. Chem. Inf. Model., vol. 61, no. 8, pp. 3891–3898, Aug. 2021, doi: 10.1021/acs.jcim.1c00203.
D.-H. Kim and S.-M. Kang, “Stapled Peptides: An Innovative and Ultimate Future Drug Offering a Highly Powerful and Potent Therapeutic Alternative,” Biomimetics, vol. 9, no. 9, p. 537, Sep. 2024, doi: 10.3390/biomimetics9090537.
Y. Li et al., “Therapeutic stapled peptides: Efficacy and molecular targets,” Pharmacological Research, vol. 203, p. 107137, May 2024, doi: 10.1016/j.phrs.2024.107137.
G. Rosenman and B. Apter, “Bioinspired materials: Physical properties governed by biological refolding,” Applied Physics Reviews, vol. 9, no. 2, p. 021303, Jun. 2022, doi: 10.1063/5.0079866.
V. I. Paat, A. A. Aloanis, and J. M. J. Najoan, “Molecular Docking Of Cyclosenegalin A As Anticancer,” Fullerene Journal of Chemistry, vol. 10, no. 1, pp. 26–33, 2025.
O. Adedirin, R. A. Abdulsalam, K. O. Nasir-Naeem, A. A. Oke, A. O. Jubril, and S. Sabiu, “Density functional theory and molecular dynamics simulation-based bioprospection of Agathosma betulina essential oil metabolites against protein tyrosine phosphatase 1B for interventive antidiabetic therapy,” Heliyon, vol. 11, no. 3, p. e42239, Feb. 2025, doi: 10.1016/j.heliyon.2025.e42239.
J. N. Martins, J. C. Lima, and N. Basílio, “Selective Recognition of Amino Acids and Peptides by Small Supramolecular Receptors,” Molecules, vol. 26, no. 1, Art. no. 1, Jan. 2021, doi: 10.3390/molecules26010106.
A. K. Oyebamiji, S. A. Akintelu, F. E. Olujinmi, O. Ebenezer, E. T. Akintayo, and C. O. Akintayo, “Design, synthesis, and pharmacological profiling of cyclic tetra-peptide derivatives for opioid receptor modulation: a review,” Discov Mol, vol. 2, no. 1, p. 12, May 2025, doi: 10.1007/s44345-025-00022-y.
E. J. Millan-Casarrubias, Y. V. García-Tejeda, C. H. González-De la Rosa, L. Ruiz-Mazón, Y. M. Hernández-Rodríguez, and O. E. Cigarroa-Mayorga, “Molecular Docking and Pharmacological In Silico Evaluation of Camptothecin and Related Ligands as Promising HER2-Targeted Therapies for Breast Cancer,” Current Issues in Molecular Biology, vol. 47, no. 3, Art. no. 3, Mar. 2025, doi: 10.3390/cimb47030193.
P. C. Agu et al., “Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management,” Sci Rep, vol. 13, p. 13398, Aug. 2023, doi: 10.1038/s41598-023-40160-2.
Copyright (c) 2025 Jessika Maya Jovanka Najoan, Rymond Jusuf Rumampuk, Vlagia Indira Paat, Anderson Arnold Aloanis

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
 This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. Copyright is retained by the authors, and articles can be freely used and distributed by others.
This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. Copyright is retained by the authors, and articles can be freely used and distributed by others.

 Jessika Maya Jovanka Najoan(1)
Jessika Maya Jovanka Najoan(1)











